Improving deep learning object detection on dental x-rays
This is a follow-up on how I improved the model from my previous post.
What was wrong:
- Precision wasn’t great
- MobileNet (SSD) performed better than Faster RCNN ResNet, which seems counter-intuitive as MobileNet is supposed to be a lighter model
- Tooth segmentation was not very precise. Often the model would output two teeth as one, and overall you could tell the model wasn’t really precise on the object boxes
In this article I will talk about two solutions I implemented to increase model accuracy:
- Image preprocessing
- Transfer learning
1. Preprocessing
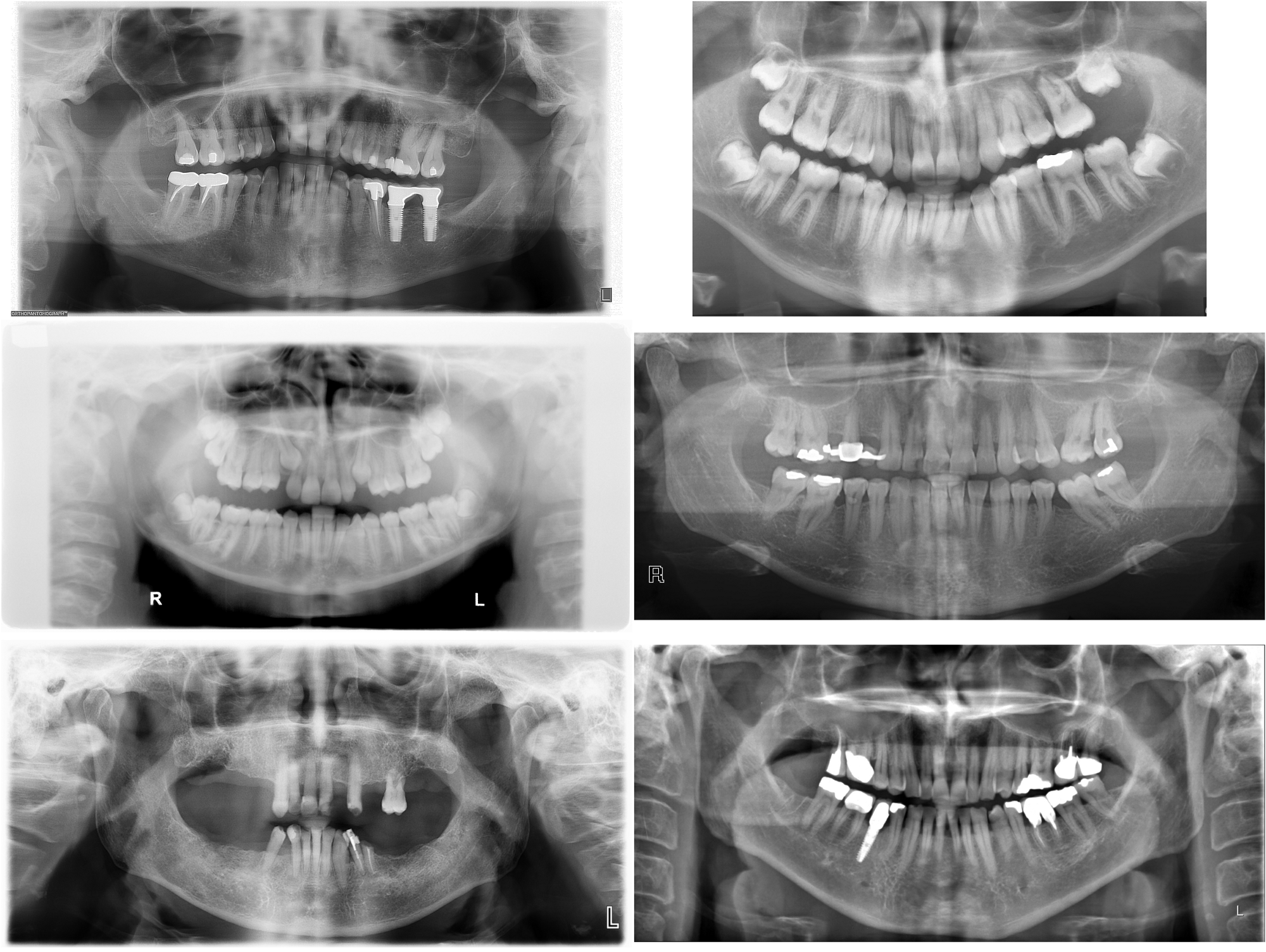
Our x-ray dataset comes from various sources, and as you can see below they vary quite a lot. There are variations in image resolution, size, contrast, and zoom on the teeth. That’s because our x-rays come from different machines used by different radiologists. Raw input sample
Image size and resolution variability is not a problem for our model because the images are all very high quality (average size 2900x1400) so it actually downsizes images before processing. Degrees of zoom on the teeth is not a problem either, it actually gives a bit of variety to our model.
However, gray level and contrast variations can make it hard for our model to really learn the right features.
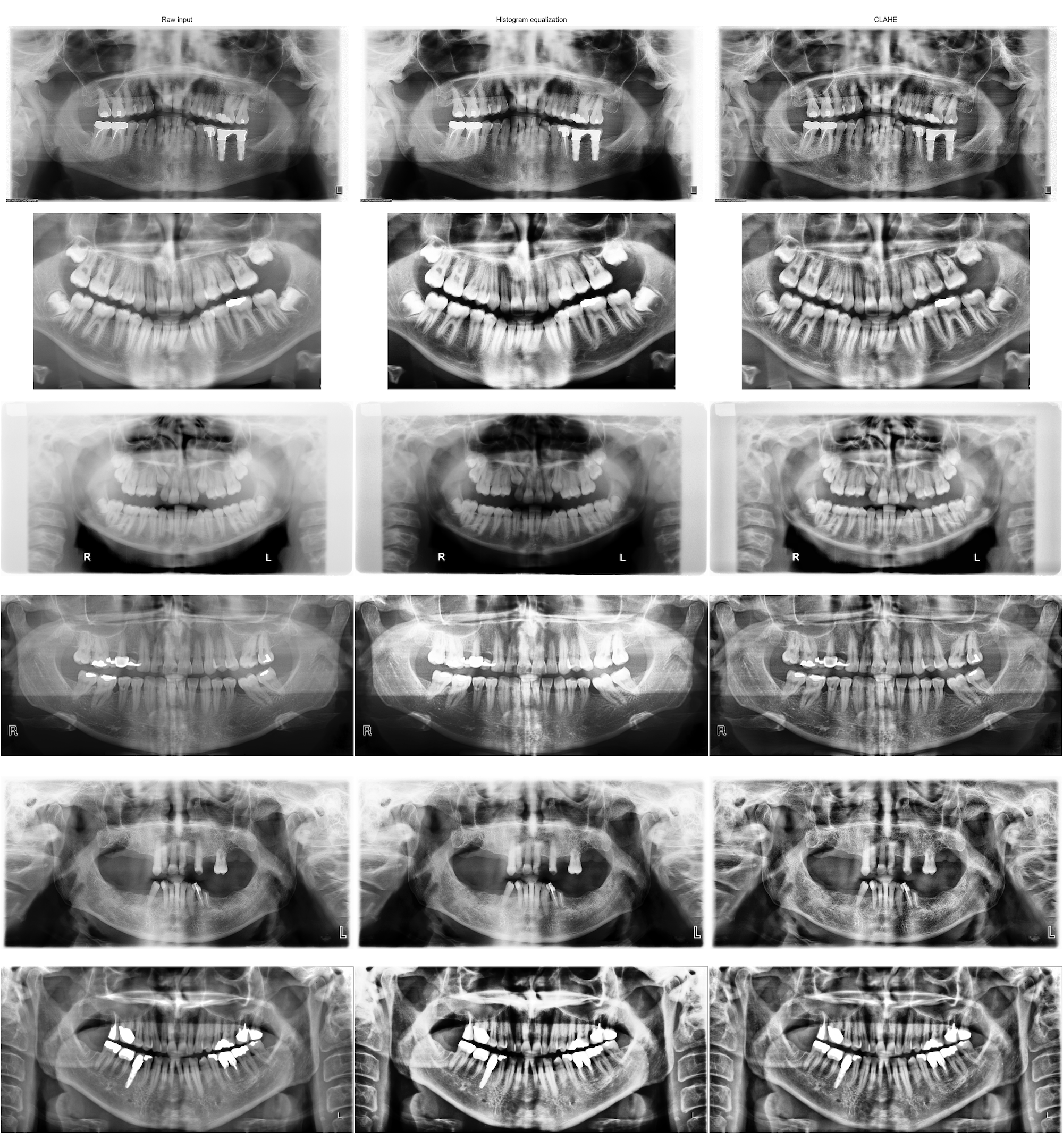
A preprocessing step would be to normalize our data to obtain images with similar contrasts and that all “look the same”. Histogram equalization is a good way to increase contrast. But for our data, simple histogram equalization is not good as it can actually reduce the visibility of tooth restorations and implants (see comparison below).
CLAHE (Contrast Limited Adaptive Histogram Equalization) is a histogram equalization technique that allows to enhance contrast locally while limiting the amplification of noise.
I used OpenCV, which is an amazing resource for editing images, and can’t recommend it enough. Here is a really nice tutorial on histogram equalization. I was able to simply use CLAHE as follow:
1
2
3
4
5
def equalize_clahe_image(image_path):
img = cv2.imread(image_path, 0)
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(16,16))
cl = clahe.apply(img)
cv2.imwrite(image_path, cl)
Empirically, a tile of (16,16) was a good tradeoff for our contrast among all images.
2. Transfer learning
Tensorflow Object Detection API makes it easy to do transfer learning from an existing model. The model zoo allows you to pick a pre-trained model and easily train it on your dataset.
I chose to use faster_rcnn_resnet50_coco for its relatively good speed and mAP score on the COCO dataset.
After 1500 iterations on my laptop, the model is already performing quite well. I used vertical and horizontal flipping for data augmentation. When comparing with my previous model, we can see how much the current model has improved:
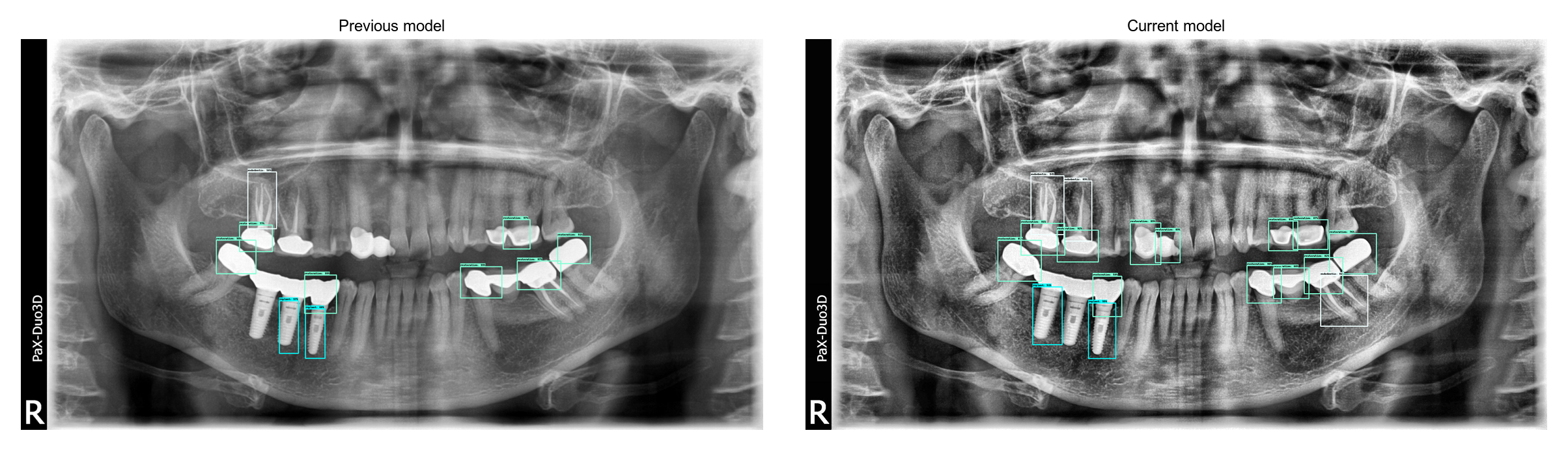
Transfer learning helped for two main reasons. First of all, a pre-trained model makes it easy to learn shapes and specific objects. Secondly, the Faster RCNN ResNet50 uses higher image sizes than my previous models, which probably helped the precision. I also had to lower the IoU threshold for non max suppression from the tensorflow config.
Current model is still not perfect, especially around implants but it is probably because we don’t have a lot of implant examples in our dataset. I haven’t trained it on GPU which limits the total amount of training possible and makes the current model only a prototype. However the jump in precision indicates that the preprocessing and transfer learning are going in the right direction.
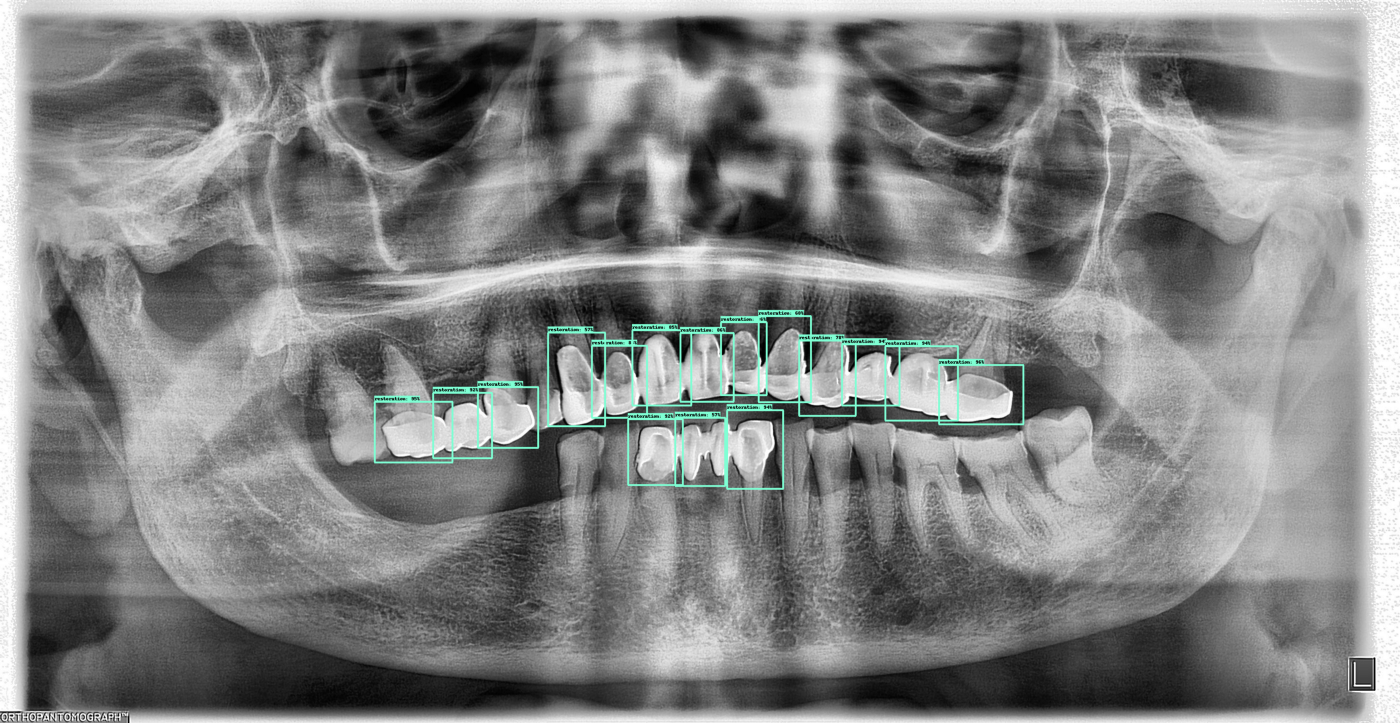
Here are a few more outputs from the current algorithm:
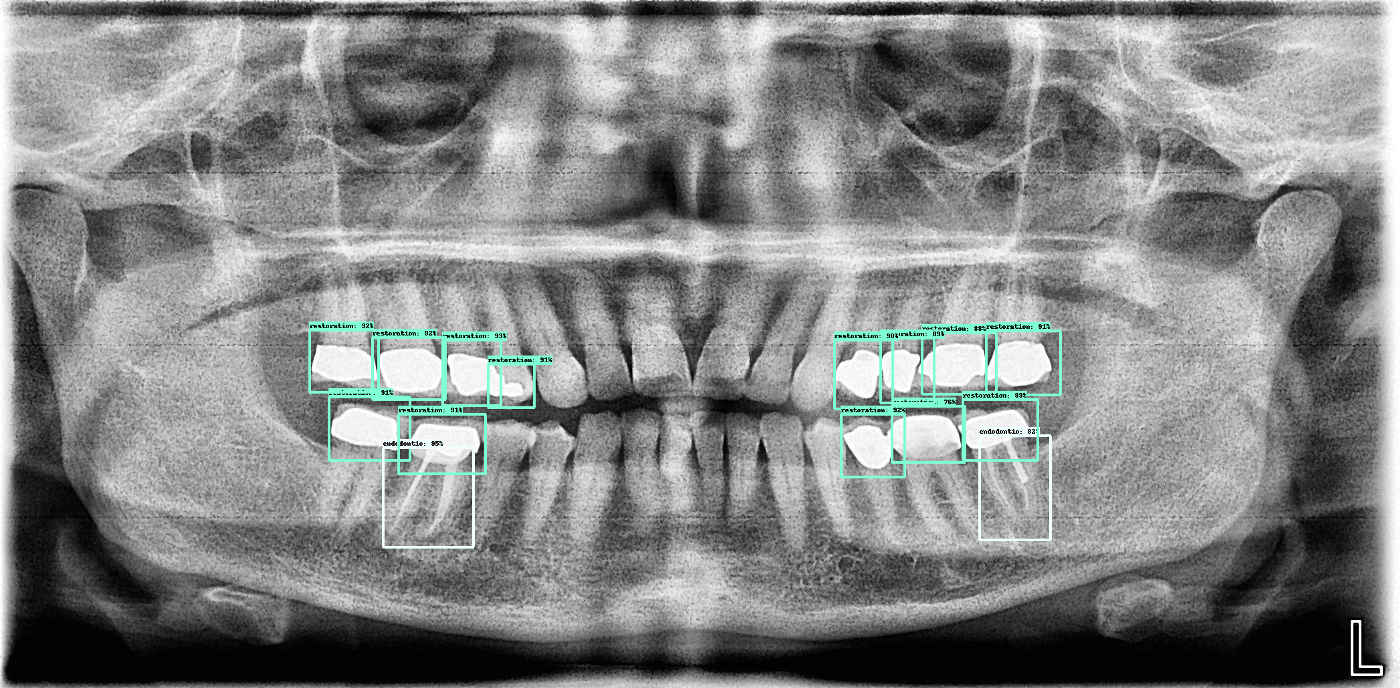
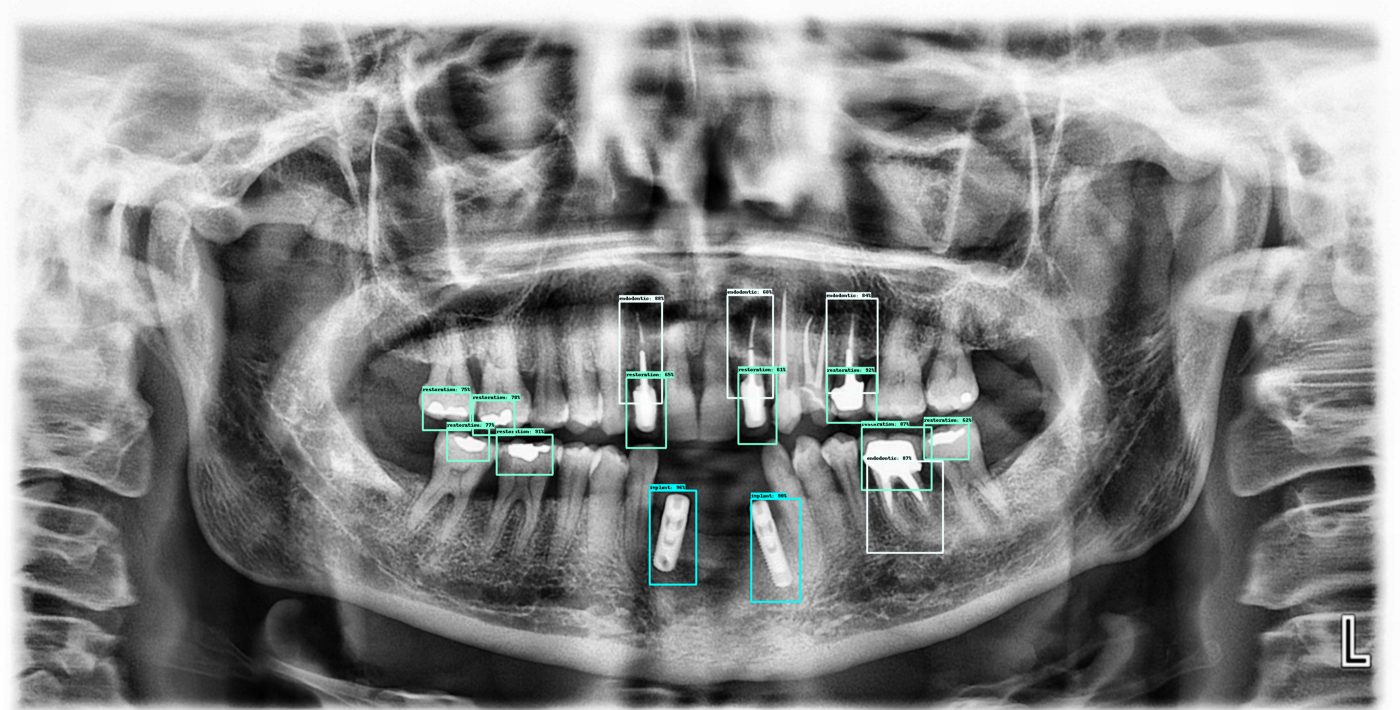
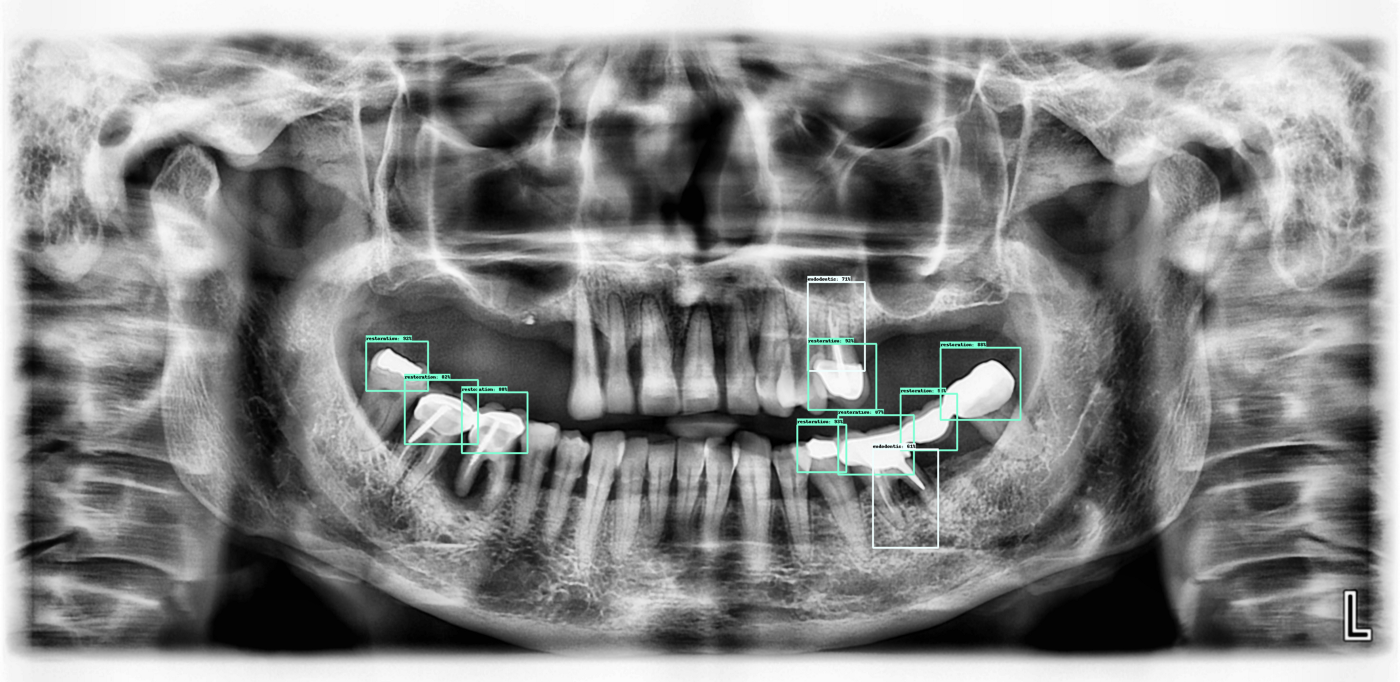
Next steps:
- Training a lot more, on GPU
- Other models (SSD, bigger ResNets…) comparisons
You can find the code here: https://github.com/clemkoa/tooth-detection
The dataset is not public yet, but we are working on it!
Disclaimer: all x-rays in the dataset have been anonymized for privacy concerns.